Unit 2: Basic Bioinformatics Tools and Resources
BIO 255: Bioinformatics and Computer Applications in Biotechnology
Why learn to manage genomic information, navigate biological databases, perform sequence analysis, and carry out comparative analysis?
The short answer: This is an area of job growth.
 Advances in DNA sequencing technology have lead to the creation of enormous data sets and databases containing a wealth of information about species of interest and individuals within a species (Humans). For example, Next Generation Sequencing (NGS) is rapidly becoming a reality in modern medicine. Many scientists predict the $1,000 genome is nearly here, and when that happens a person's genome will be a common feature of their medical file. The industry will need Lab Technicians to run the assays, Bioinformatics Technicians to process the data, Health Informatics Analysts to manage the wealth of patient data, and the list goes on. Each of the people filling these jobs needs an understanding of how the data is produced, managed, and potentially applied in the real world. Below you will find two articles to read. These articles were selected to give you a taste of how data is produced and how it will potentially be used. (QUIZ IN BB)
Advances in DNA sequencing technology have lead to the creation of enormous data sets and databases containing a wealth of information about species of interest and individuals within a species (Humans). For example, Next Generation Sequencing (NGS) is rapidly becoming a reality in modern medicine. Many scientists predict the $1,000 genome is nearly here, and when that happens a person's genome will be a common feature of their medical file. The industry will need Lab Technicians to run the assays, Bioinformatics Technicians to process the data, Health Informatics Analysts to manage the wealth of patient data, and the list goes on. Each of the people filling these jobs needs an understanding of how the data is produced, managed, and potentially applied in the real world. Below you will find two articles to read. These articles were selected to give you a taste of how data is produced and how it will potentially be used. (QUIZ IN BB)
-
Cost of Gene Sequencing Falls, Raising Hopes for Medical Advances - NY Times
-
High-throughput sequencing for biology and medicine -- Review article
New to Bioinformatics, Sequence Analysis, Computational Biology?
Bioinformatics is an interdisciplinary field that creates and refines methods for generating, storing and managing, and analyzing biological data. People enter this field of study/work from many backgrounds, inlcuding genetics, molecular biology, computer science, physics, and mathematics. Therefore, teams of Bioinformaticists use priniciples from engineering, math, physics, and computer science to address and process biological data. Read the chapter "Introduction to Bioinformatics" from the book Bioinformatic Technologies. This chapter offers a brief overview of bioinformatics, the tools and databases used during analysis, and some common applications of bioinformatics. Additionally, read the chapter "Genomics and Potential Downstream Applications in the Developing World" from the book Genomics Applications for the Developing World to gain more insight into the potential applications of bioinformatics and computational biology in the developing world. Keep in mind that although this chapter focuses on the developing world, the themes presented are applicable world-wide. The book chapters listed below from Genomics Applications for the Developing World describe very interesting and medically significant fields of science that capitalize on bioinformatics and computationl biology methods. These book chapters may help guide your article selection in Assignment 2.1.
-
Malaria Genomics and the Developing World
-
The Genomics of Cholera
Assignmnet 2.1-- Gene Identification
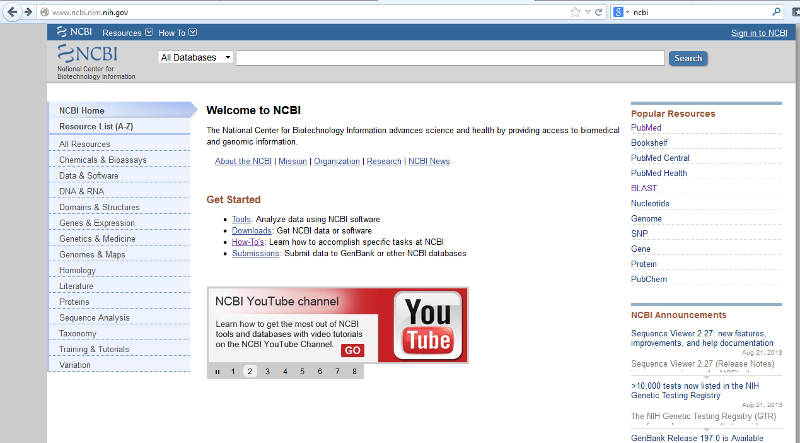
This assignment requires you to access and navigate PubMed, a database of research articles found on the National Center for Biotechnology Information (NCBI) website, and several other programs accessed through NCBI . NCBI has a YouTube channel (http://www.youtube.com/user/NCBINLM) that is useful for learning how to navigate and best utalize the many bioinformatics tools available. Notice the "PubMed" link under Popular Resources on the right side of the screen shot below.
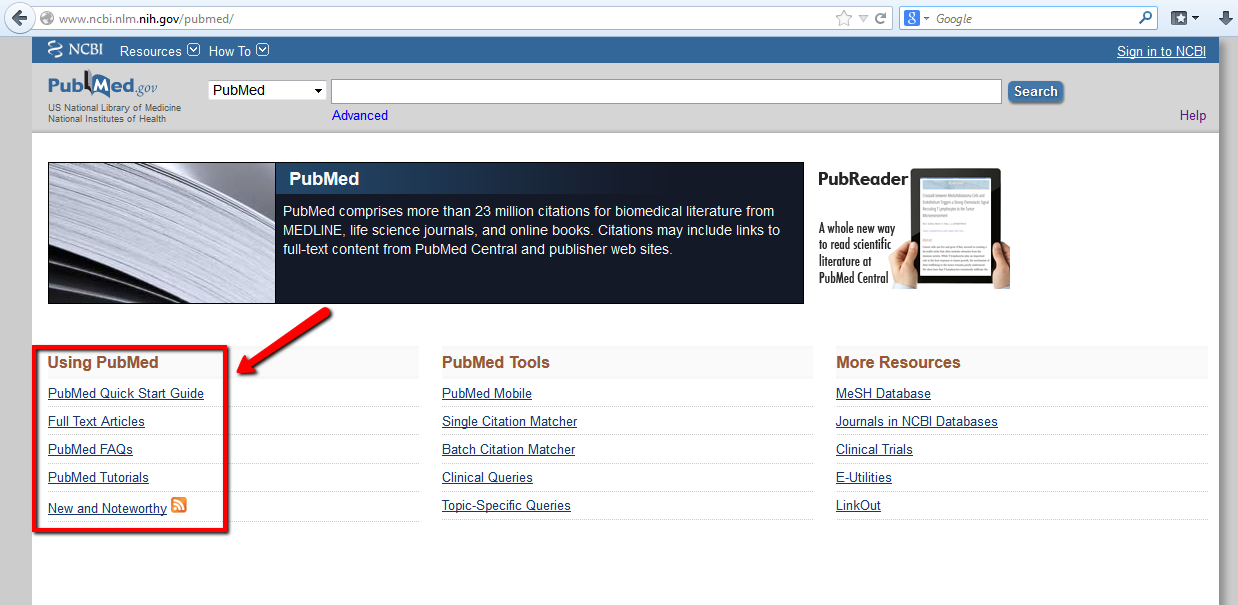
Additionally, each Database in NCBI has useful links to guide you in effective use of the resources. Follow these links for frequently asked questions (FAQs), Tutorials, and Quick Start Guides).
Select an area of Biotechnology (Health/Medicine, Industrial, Agriculture, or Environmental) and find a peer reviewed research article published no earlier than 2012 that describes at least one gene of interest. Submit your article and gene of interest (submit the gene identifier or acession number) to your instructor for approval before moving on to the next part of the assignment. Once the article and gene have been approved, write a one paragraph summaryof the article in your own words. Include a statement describing why you selected your gene of interest. In this summary you also need to provide the PubMed ID for the article you chose.
Use the appropriate NCBI database to obtain the protein and nucleotide coding sequence (report the sequence that encodes the mature mRNA sequence). Remember that eukaryotic genes almost always contain introns. You will create two text files:
(1) your DNA sequence in FASTA format and
(2) your protein sequence in FASTA format.
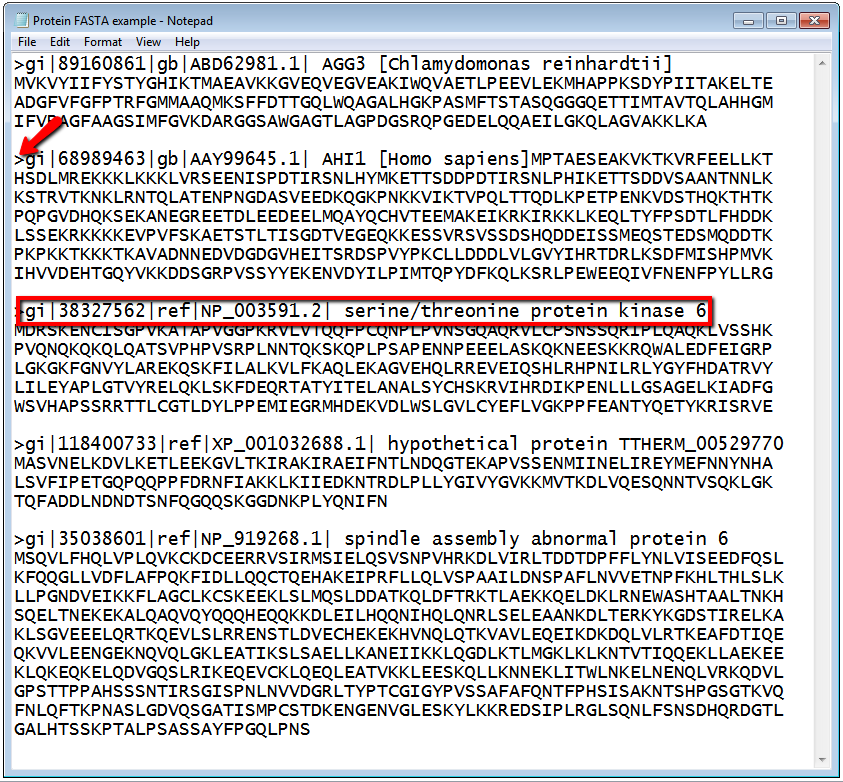
See the description of FASTA format on the NCBI website: http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml. Your initial files should look similar to the screen shot below, but with a single sequence in each file. You will soon populate your file with more sequences. In the screen shot below the Arrow is pointing to the ">" that signals a new sequence, beginning with a line of description. The red box is the identifying text or description of the sequence below. This description text needs to be unique for each sequence. The following lines are the protein or DNA sequence of interest. Text files such as these are generated routinely for use in bioinformatics programs.
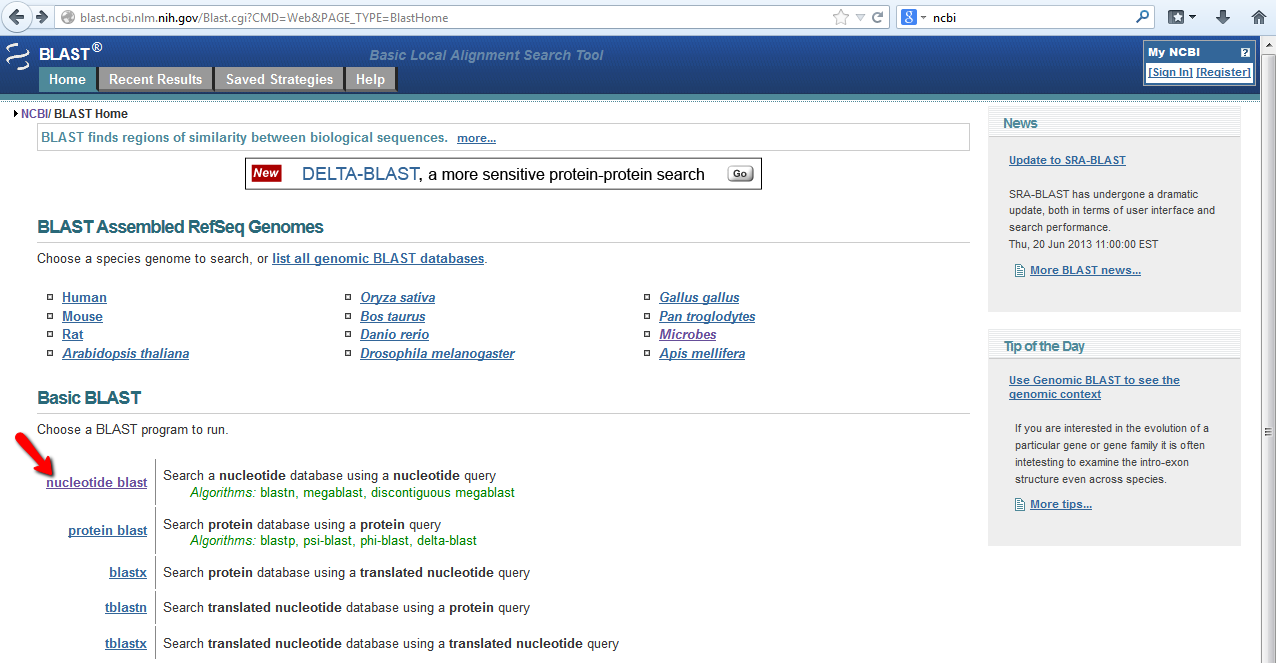
Once you have you text file with your DNA sequence of interest in FASTA format, the next step is to determine if other organisms have similar genes. To query other species for your gene of interest you will use the program BLAST found on the NCBI website. Links to BLAST tutorials and FAQs pages can be found here: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs. Remember YouTube tutorials are available. For a standard BLAST search using a DNA sequence as your inout you want to begin your search using the Standard Nucleotide BLAST, also called BLASTn in some cases, indicated by the red arrow below.
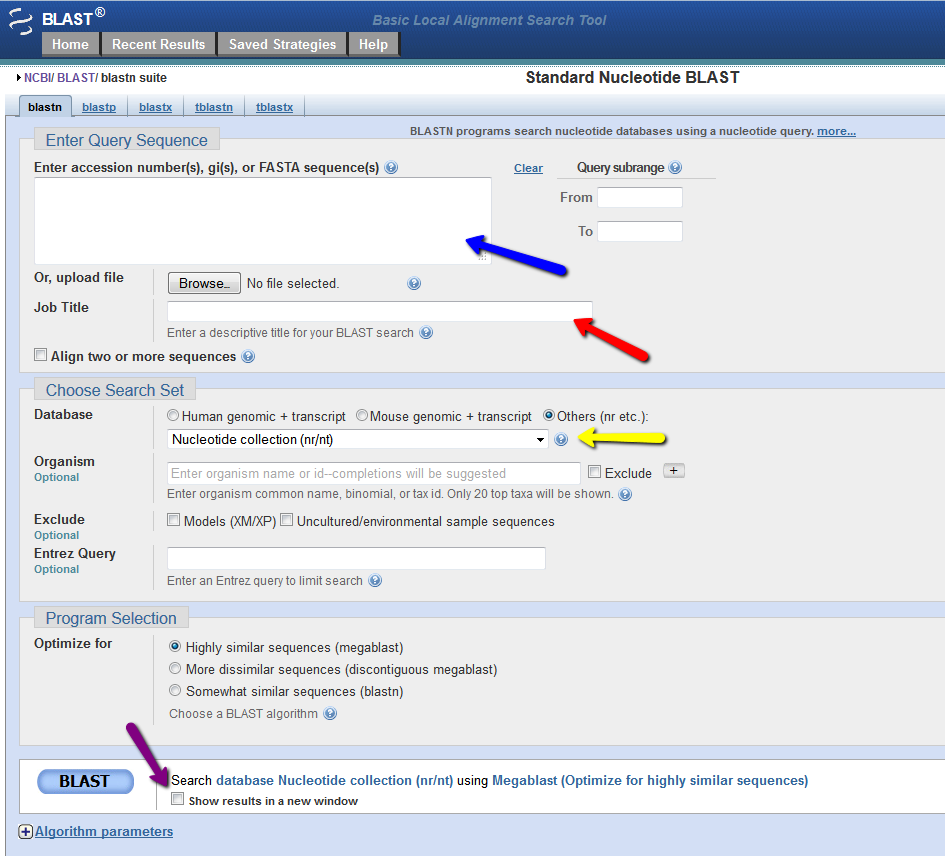
On the Standard Nucleotide BLAST page you have the option to enter your FAST sequence(s) or the accession number(s) for your gene(s) of interest (see blue arrow below). The nice thing is that you can search a database for multiple genes at one time and receive a separate report for each gene. It is a good idea to name your searches in teh "Job Title" field (red arrow below). The next you step is to select the Database you wish to query. You can obtain a description of each database by clicking the "?" (see yellow arrow below) and scrolling through the available databse options. Be careful not to select a protein database when using a nucleotide sequence for your Standard Nucleotide BLAST. For people who wish to use more advanced settings, you can alter the "Algorithm parameters" found at the bottom of the screen (see screen shot below). This extra section will allow you to set the "Max target sequences" and alter "Scoring Parameters". Finally, before you click the big "BLAST" button, you may want to select the "Show results in a new window" option, which will allow you to more easily run multiple searches at once (see purple arrow below). If your search does not yeild sequence hits from more than 5 species then you may want to change your Program select to "More dissimilar sequences (discontiguous megablast)" (see screen shot below).
When you web browser refreshes with your results there are several key sections you will want to analyze. You are encouraged to take and save screen shots similar to the four "BLAST results Screen Shots" below to use in Assignemnt 2.3, where you will make a Comparative Genomics Tutorial. At the top of your BLAST reulsts screen you will find the descriptive information for your search, inculding the program used, the length of your query sequence, the type of molecule used in the search (protein or nucleotide), and the database youy searched (See BLAST results Screen Shot 1). Following this desciptive text you will see the Graphic Summary of your search results (BLAST results Screen Shot 2). This display shows you at a glance how well the "Hits" line up with your query sequence. As indicated in the screen shot you can mouse over each colored bar and click to jump down to the alignment of your sequence with the specific BLAST hit. The third section of your results page lists a brief description of each Hit. The decription lines include the species name, gene name, and importantly the link to the full sequence. The other value to pay attention to is the E-value. This number indicates the "number of hits one can "expect" by chance when searching a database of a particular size", as described in the BLAST FAQs. The closer this value is to 0 (zero) the better the hit, and the list is ordered from best hit to lowest scoring hit. The final section of the results screen shows the alignments between your query sequence and the hit. Some panels in this section may show multiple alignments, since the sequences may line up with the query and some sequences will only align in certain areas. To complete your DNA sequence FASTA file select the top ten (10) sequences from from at least five (5) different species listed in your BLAST results, not including the hit for the query sequence. Obtain the nucleotide sequences and list them in FASTA format under your original gene of interest. Repeat this process for your protein sequence, but instead of using the Standard Nulceotide BLAST, use the Standard Protein BLAST. In the end you should have two text files listing (1) eleven DNA sequences in FASTA format, and (2) eleven protein sequences in FASTA format. Using the data generated and listed in your two files, use a word processor program to create a document that includes a table listing the top ten nucleotide and protein hits in a clear manner. Under this table you write a brief description of the similarities and difference between the nucleotide and protein BLAST search results. This summary will include which datbase was quried for each search strategy, what style of input (sequence or gene identifier) was used, and number of hits obtained with each search strategy.
BLAST results Screen Shot 1
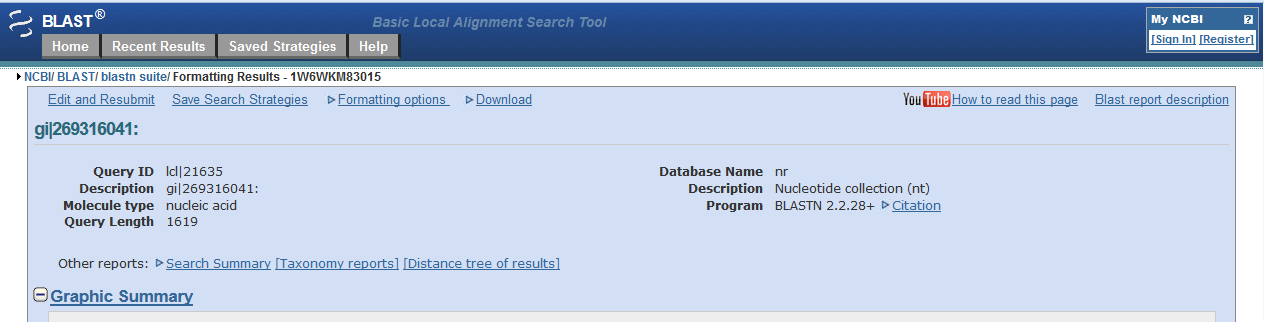
BLAST results Screen Shot 2
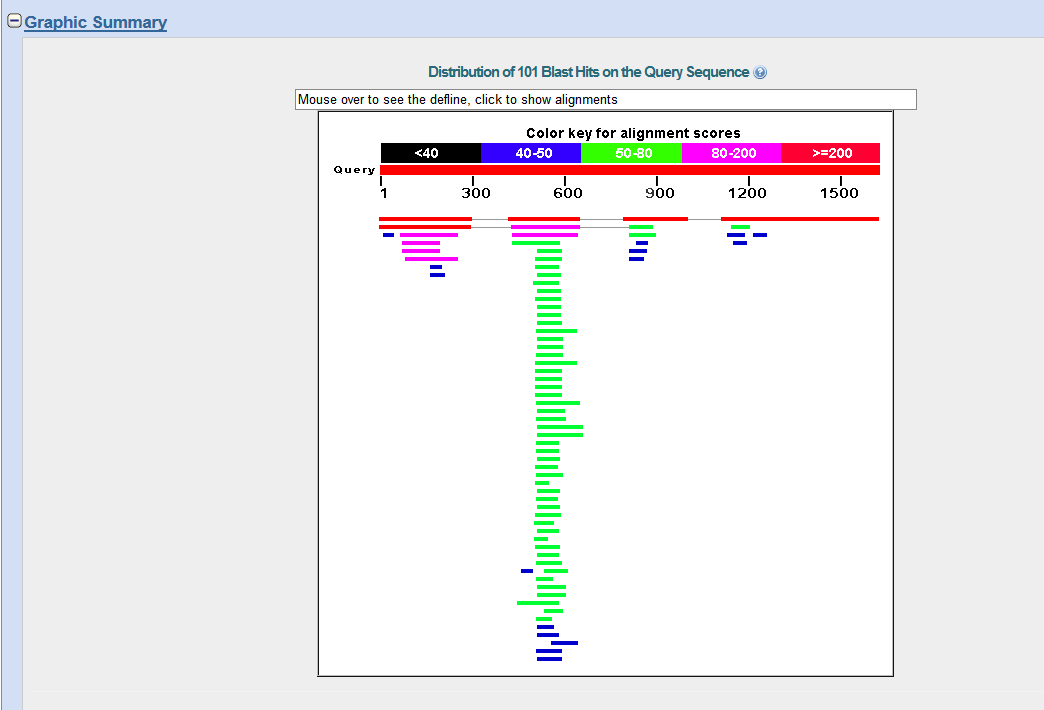
BLAST results Screen Shot 3
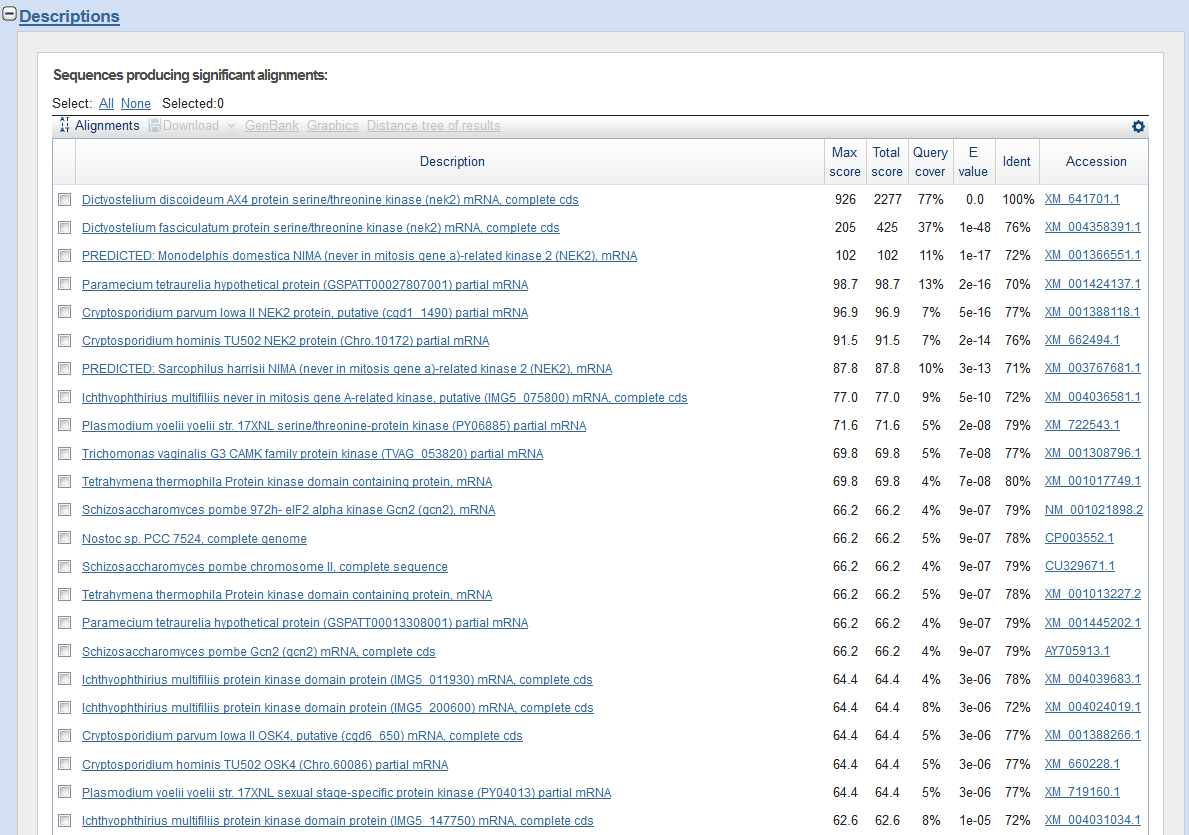
BLAST results Screen Shot 4
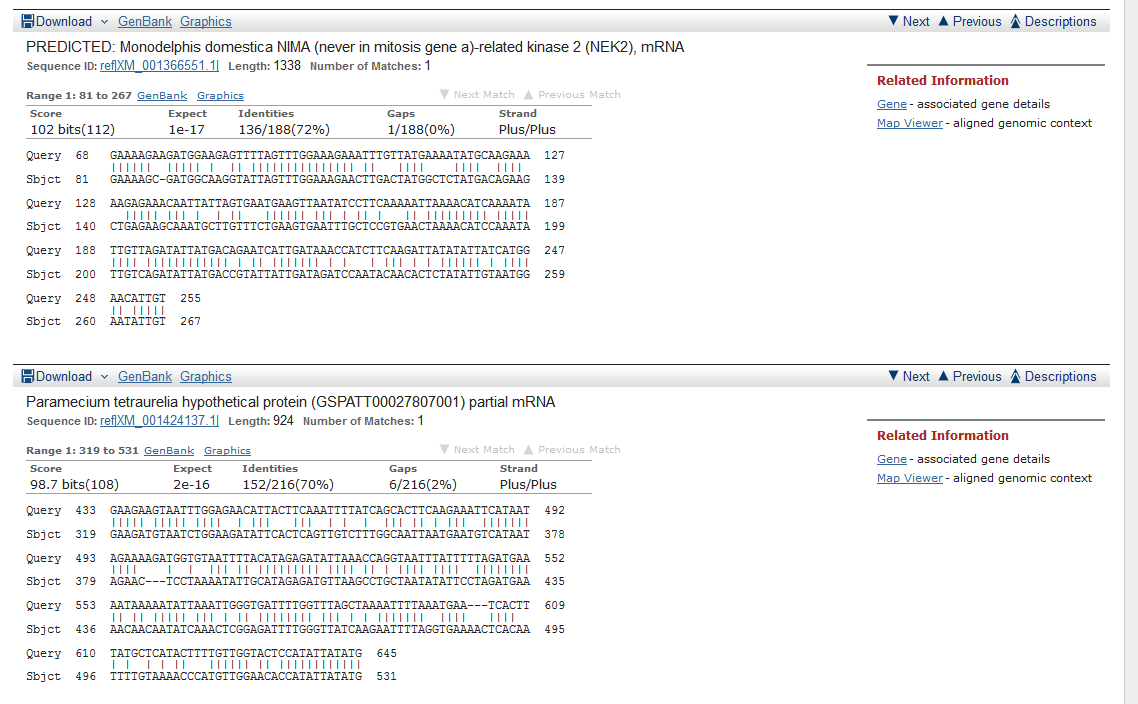
Assignment 2.2 - Introduction to Sequence Analysis
Obtaining DNA and protein sequences of interest is only the beginning of sequence analysis. Some common sequence information that you may need to know about your sequences include determining how closely realted two sequences are two each other, calculating the G-C content of nucleotide sequences, translation of nucleotide sequences, and discovering conserved regions among a related collection of protein sequences. Assignment 2.2 directs you to begin analyzing the sequences in your DNA and protein files created in Assignment 2.1. Read the chapter "What is Comparative Genomics" from the book Comparative Genomics for a more detailed description of how sequence analysis is used to answer significant biological questions.
Your analysis steps (Be sure to tabulate and organize your data as you generate it):
-
Estimate the percent identity between your original gene of interest and the 10 other sequences in your DNA FASTA file. Begin by using the LAlign program found here: http://embnet.vital-it.ch/software/LALIGN_form.html. The LAlign program is a basic program designed to align your two sequences, output the alignment, and report the percent identity between the two submitted sequences. You want to set the program to use a "global alignment", otherwise it will default to giving you several partial alignments. Explore online and check out other programs that you may like better. Another useful program that gives both percent identity and similarity can be found here http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::stretcher and is called EMBOSS 6.3.1: stretcher. It is possible to use a local (on your computer) program to determine the percent identity between sequences. The BioEdit program, described below can open your FASTA files and from the Menu you can select Sequence --> Pairwise Alignment --> Align Two sequences (optimal Glabal alignment). See the instruction video below for details.

Using the BioEdit Program Video Tutorial
-
Use a web-based tool calculate the molecular weight (in Daltons, Da) and the isoelectric point (pI) of each protein in your Protein FASTA file. The pI of a protein is the pH required to maintain a molecule at no net electrical charge. In other words, the protein's negative and positive charges are equal. This information is useful for designing protein biochemistry and column chromatography experiments.
-
Using your Protein FASTA file perform a multiple alignment of your protein sequences using the Clustal Omega program found on the ExPASy website. To visualize and analyze your output alignment file you will need to download and install either BioEdit (PC - http://www.mbio.ncsu.edu/bioedit/page2.html) or Sea View (Mac - http://pbil.univ-lyon1.fr/software/seaview.html). Using BioEdit calculate the percent identity and similarity between your original protein of interest and the 10 other protein sequences in the multiple alignment. Next use BioEdit to generate a nice graphic image of your alignment. If you are using a MAC and will visualize your alignment with Sea View then open your alignment file in the program and use the generate .pdf function under the File menu to create an image of your alignment. Using Jing or another screen capture program take a snap shot of as much of the alignement as you can to include in your data analysis summary. For MAC users, use programs like LAlign to determine the percent identity between your protein of interest and other sequences in your Protein FASTA file. Unfortunaltely, BioEdit is not available for Macs, but for MAC users Sea View has some additional functionalities that BioEdit lacks. Definetely explore the potential of this program, using your DNA and Protein FASTA files as test cases. Videos illustrating how to navigate the the Clustal Omega program and BioEdit are below.
-
Compile all of your generated data, inlcuding a color image of your protein multiple alignment, in a spreadsheet program. Pay attention to the presentation and ease of access to your data. Make use of formatting learned in Unit 1 to aid in your organization and presentation of your data. Finally be sure to indicate the web-based program used to generate the data you present in your spreadsheet.
Assignment 2.3 - Comparative Genomics Tutorial
Part of your job as a laboratory professional will be to teach incoming lab personnel the necessary tools to be successful. You will create a tutorial using a word processor that outlines your search stragey and sequence analysis in a clear and approachable manner. This tutorial is to be designed for an incoming Biotechnology Student who is new to bioinformatics. You should include information students will need to identify genes and proteins and complete basic sequence analysis. Use your sequence of interest identified in Assignment 2.1 as the example throughout your tutorial. Be sure to describe the databases used, basic functionality of the databases and the infromation obtained from potential searches. You are advised to use screen capture tools such as Jing to illustrate steps. This tutorial will be posted on the discussion board and you are required to meaningfully comment on all your peers' tutorials.
 Advances in DNA sequencing technology have lead to the creation of enormous data sets and databases containing a wealth of information about species of interest and individuals within a species (Humans). For example, Next Generation Sequencing (NGS) is rapidly becoming a reality in modern medicine. Many scientists predict the $1,000 genome is nearly here, and when that happens a person's genome will be a common feature of their medical file. The industry will need Lab Technicians to run the assays, Bioinformatics Technicians to process the data, Health Informatics Analysts to manage the wealth of patient data, and the list goes on. Each of the people filling these jobs needs an understanding of how the data is produced, managed, and potentially applied in the real world. Below you will find two articles to read. These articles were selected to give you a taste of how data is produced and how it will potentially be used. (QUIZ IN BB)
Advances in DNA sequencing technology have lead to the creation of enormous data sets and databases containing a wealth of information about species of interest and individuals within a species (Humans). For example, Next Generation Sequencing (NGS) is rapidly becoming a reality in modern medicine. Many scientists predict the $1,000 genome is nearly here, and when that happens a person's genome will be a common feature of their medical file. The industry will need Lab Technicians to run the assays, Bioinformatics Technicians to process the data, Health Informatics Analysts to manage the wealth of patient data, and the list goes on. Each of the people filling these jobs needs an understanding of how the data is produced, managed, and potentially applied in the real world. Below you will find two articles to read. These articles were selected to give you a taste of how data is produced and how it will potentially be used. (QUIZ IN BB)