Assignmnet 2.1-- Gene Identification
This assignment requires you to access and navigate PubMed, a database of research articles found on the National Center for Biotechnology Information (NCBI) website, and several other programs accessed through NCBI . NCBI has a YouTube channel (http://www.youtube.com/user/NCBINLM) that is useful for learning how to navigate and best utalize the many bioinformatics tools available. Notice the "PubMed" link under Popular Resources on the right side of the screen shot below.
Additionally, each Database in NCBI has useful links to guide you in effective use of the resources. Follow these links for frequently asked questions (FAQs), Tutorials, and Quick Start Guides).
Select an area of Biotechnology (Health/Medicine, Industrial, Agriculture, or Environmental) and find a peer reviewed research article published no earlier than 2012 that describes at least one gene of interest. Submit your article and gene of interest (submit the gene identifier or acession number) to your instructor for approval before moving on to the next part of the assignment. Once the article and gene have been approved, write a one paragraph summaryof the article in your own words. Include a statement describing why you selected your gene of interest. In this summary you also need to provide the PubMed ID for the article you chose.
Use the appropriate NCBI database to obtain the protein and nucleotide coding sequence (report the sequence that encodes the mature mRNA sequence). Remember that eukaryotic genes almost always contain introns. You will create two text files:
(1) your DNA sequence in FASTA format and
(2) your protein sequence in FASTA format.
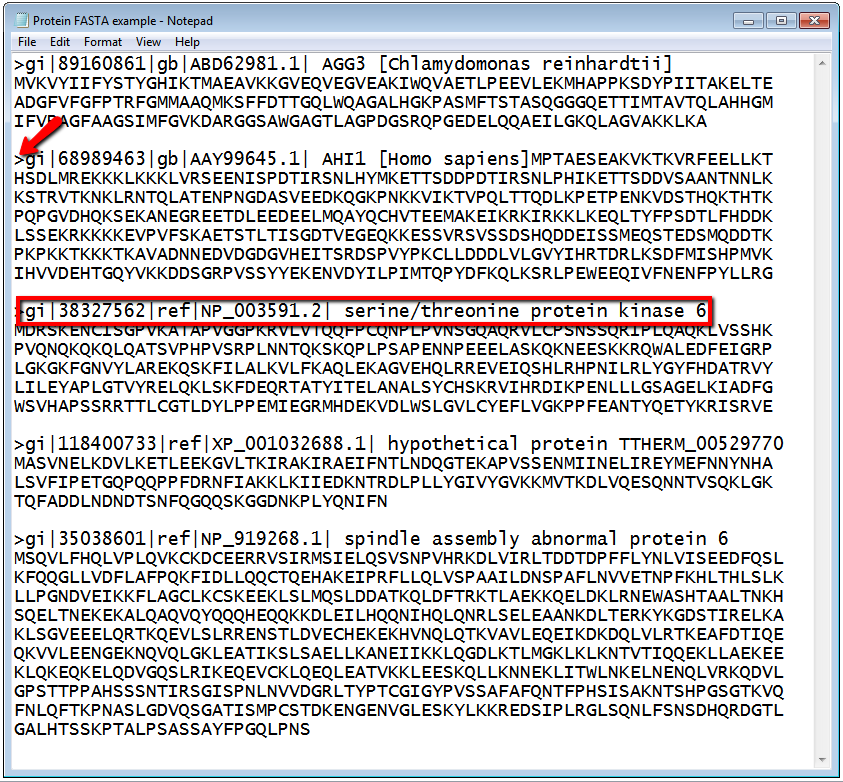
See the description of FASTA format on the NCBI website: http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml. Your initial files should look similar to the screen shot below, but with a single sequence in each file. You will soon populate your file with more sequences. In the screen shot below the Arrow is pointing to the ">" that signals a new sequence, beginning with a line of description. The red box is the identifying text or description of the sequence below. This description text needs to be unique for each sequence. The following lines are the protein or DNA sequence of interest. Text files such as these are generated routinely for use in bioinformatics programs.
Once you have you text file with your DNA sequence of interest in FASTA format, the next step is to determine if other organisms have similar genes. To query other species for your gene of interest you will use the program BLAST found on the NCBI website. Links to BLAST tutorials and FAQs pages can be found here: http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs. Remember YouTube tutorials are available. For a standard BLAST search using a DNA sequence as your inout you want to begin your search using the Standard Nucleotide BLAST, also called BLASTn in some cases, indicated by the red arrow below.
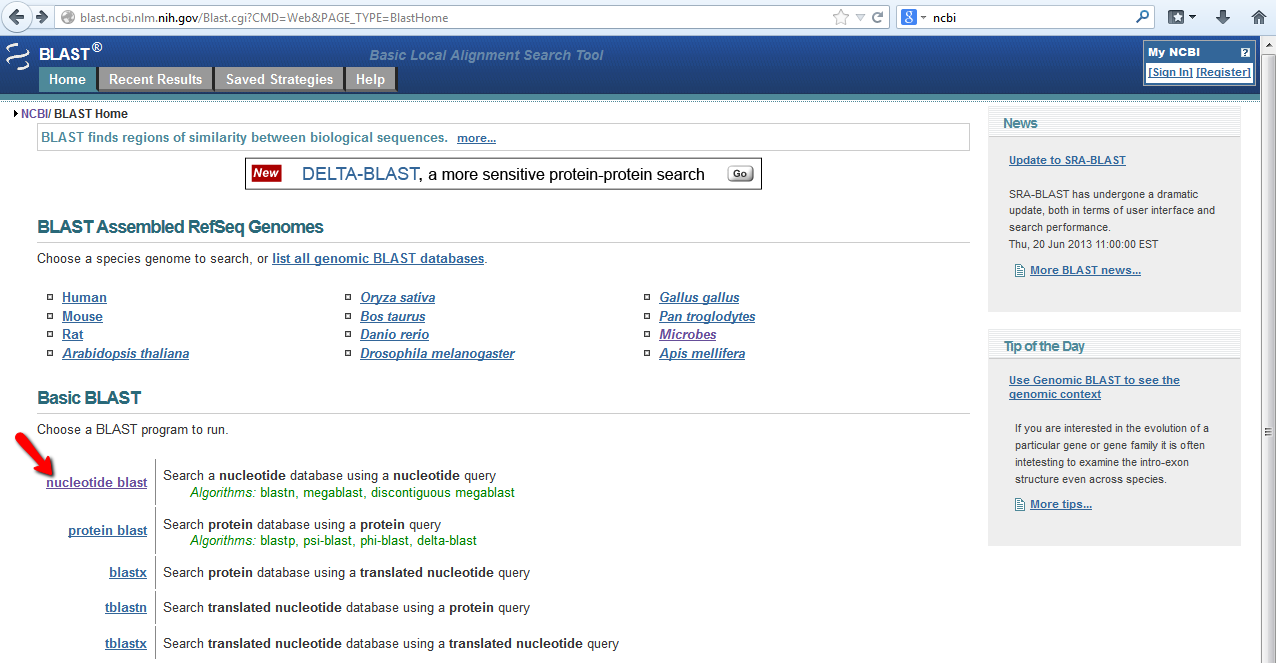
On the Standard Nucleotide BLAST page you have the option to enter your FAST sequence(s) or the accession number(s) for your gene(s) of interest (see blue arrow below). The nice thing is that you can search a database for multiple genes at one time and receive a separate report for each gene. It is a good idea to name your searches in teh "Job Title" field (red arrow below). The next you step is to select the Database you wish to query. You can obtain a description of each database by clicking the "?" (see yellow arrow below) and scrolling through the available databse options. Be careful not to select a protein database when using a nucleotide sequence for your Standard Nucleotide BLAST. For people who wish to use more advanced settings, you can alter the "Algorithm parameters" found at the bottom of the screen (see screen shot below). This extra section will allow you to set the "Max target sequences" and alter "Scoring Parameters". Finally, before you click the big "BLAST" button, you may want to select the "Show results in a new window" option, which will allow you to more easily run multiple searches at once (see purple arrow below). If your search does not yeild sequence hits from more than 5 species then you may want to change your Program select to "More dissimilar sequences (discontiguous megablast)" (see screen shot below).
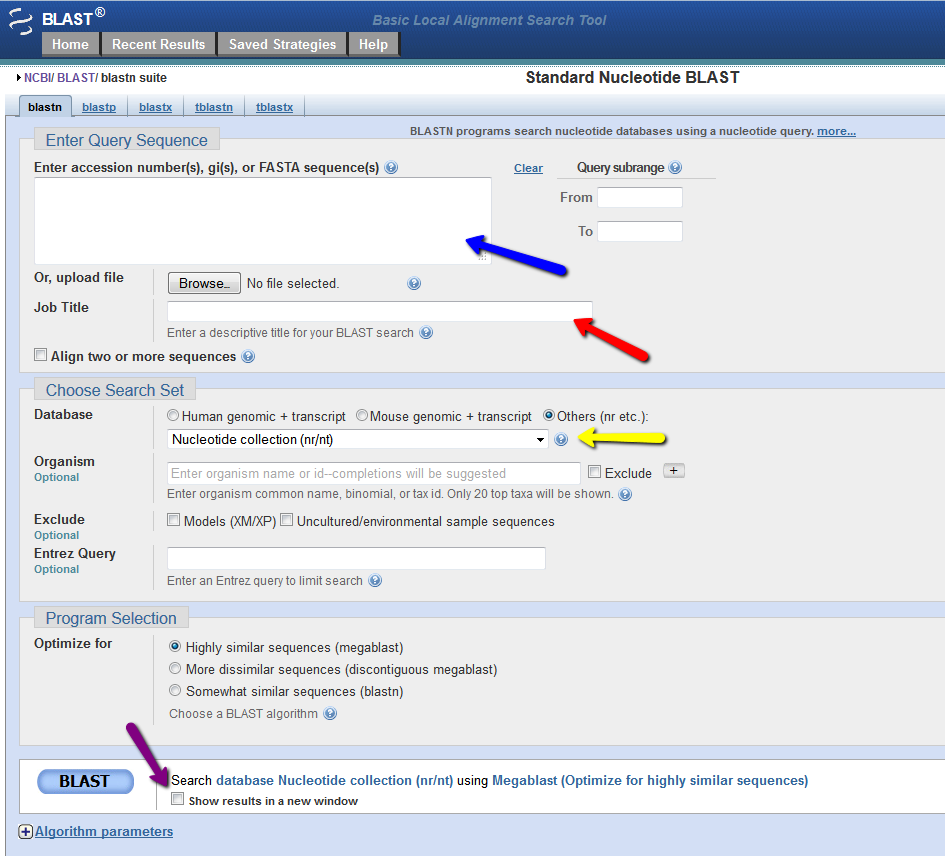
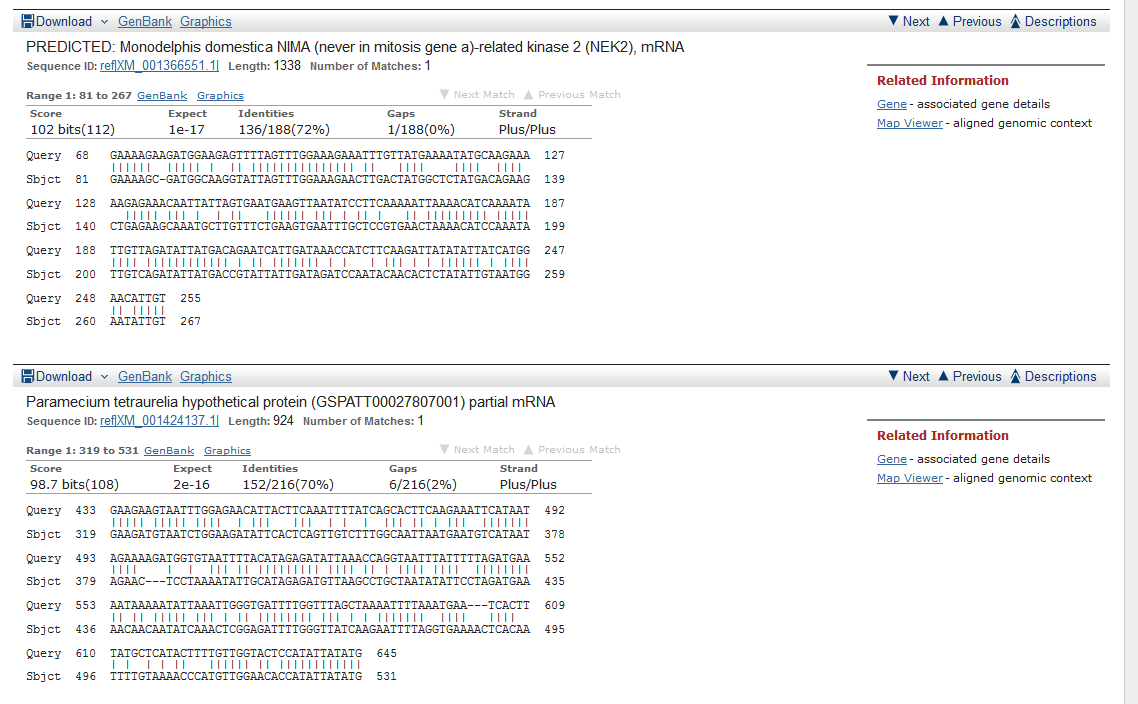
When you web browser refreshes with your results there are several key sections you will want to analyze. You are encouraged to take and save screen shots similar to the four "BLAST results Screen Shots" below to use in Assignemnt 2.3, where you will make a Comparative Genomics Tutorial. At the top of your BLAST reulsts screen you will find the descriptive information for your search, inculding the program used, the length of your query sequence, the type of molecule used in the search (protein or nucleotide), and the database youy searched (See BLAST results Screen Shot 1). Following this desciptive text you will see the Graphic Summary of your search results (BLAST results Screen Shot 2). This display shows you at a glance how well the "Hits" line up with your query sequence. As indicated in the screen shot you can mouse over each colored bar and click to jump down to the alignment of your sequence with the specific BLAST hit. The third section of your results page lists a brief description of each Hit. The decription lines include the species name, gene name, and importantly the link to the full sequence. The other value to pay attention to is the E-value. This number indicates the "number of hits one can "expect" by chance when searching a database of a particular size", as described in the BLAST FAQs. The closer this value is to 0 (zero) the better the hit, and the list is ordered from best hit to lowest scoring hit. The final section of the results screen shows the alignments between your query sequence and the hit. Some panels in this section may show multiple alignments, since the sequences may line up with the query and some sequences will only align in certain areas. To complete your DNA sequence FASTA file select the top ten (10) sequences from from at least five (5) different species listed in your BLAST results, not including the hit for the query sequence. Obtain the nucleotide sequences and list them in FASTA format under your original gene of interest. Repeat this process for your protein sequence, but instead of using the Standard Nulceotide BLAST, use the Standard Protein BLAST. In the end you should have two text files listing (1) eleven DNA sequences in FASTA format, and (2) eleven protein sequences in FASTA format. Using the data generated and listed in your two files, use a word processor program to create a document that includes a table listing the top ten nucleotide and protein hits in a clear manner. Under this table you write a brief description of the similarities and difference between the nucleotide and protein BLAST search results. This summary will include which datbase was quried for each search strategy, what style of input (sequence or gene identifier) was used, and number of hits obtained with each search strategy.
BLAST results Screen Shot 1
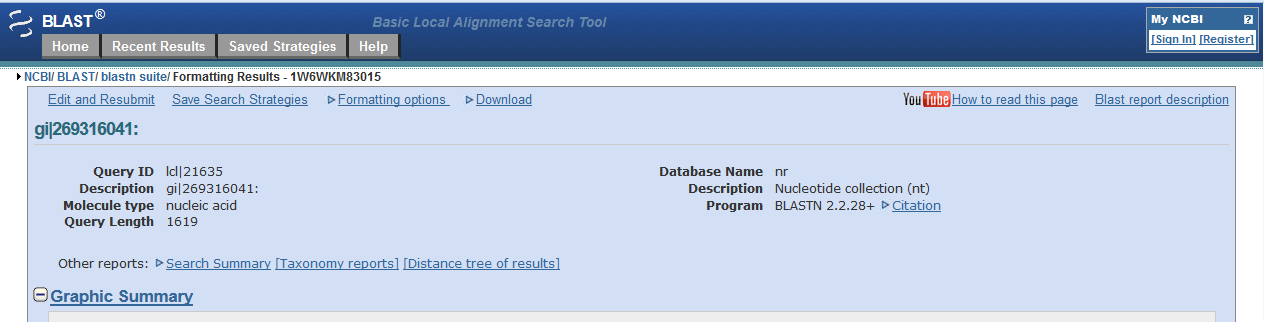
BLAST results Screen Shot 2
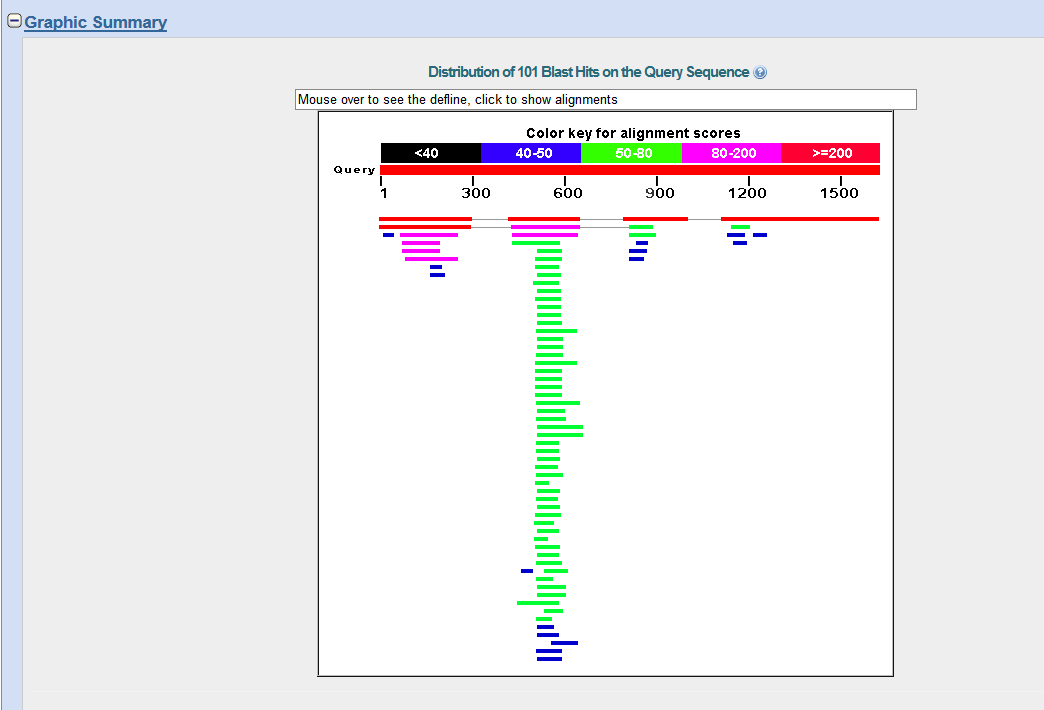
BLAST results Screen Shot 3
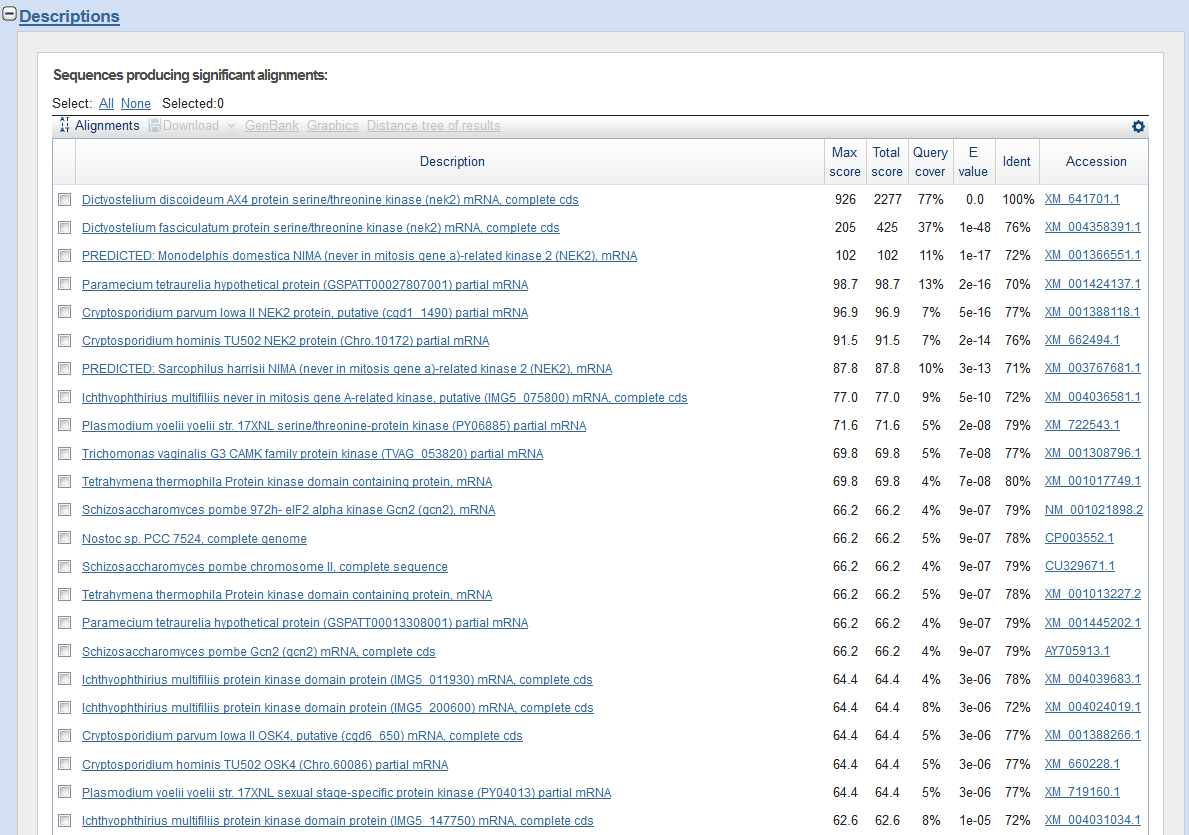
BLAST results Screen Shot 4